Now, what was missing until today, was easy access to gists in Bioclipse itself. No
gist.load(33421) yet. There still is not, but I uploaded earlier today a Wizard for it. (The manager will follow later). Right click on an open Project, select New -> Other, and pick Download Gist: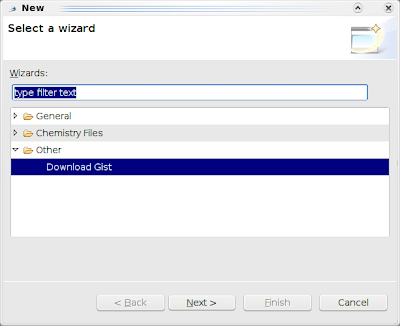 and click Next:
and click Next: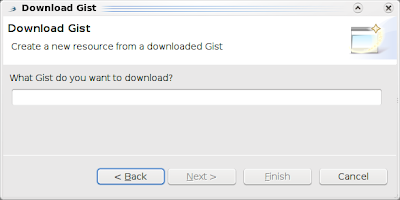 Then, just type the number of the Gist you want to open in Bioclipse, for example 18315 (see Bioclipse2 Scripting #1: from SMILES to a UFF optimized structure in Jmol), and click another Next to select a file name and location:
Then, just type the number of the Gist you want to open in Bioclipse, for example 18315 (see Bioclipse2 Scripting #1: from SMILES to a UFF optimized structure in Jmol), and click another Next to select a file name and location: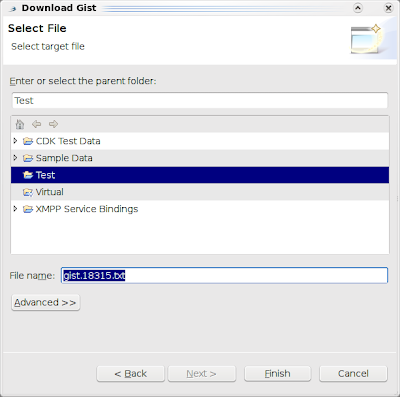 The current code does require you to know the Gist number, so you'll need a web browser to look it up, but we do have search facilities in mind. Also, while the code attempts so, the resulting Gist is not automatically openend in an editor (a bug). Another idea is to just install the egit plugin in Bioclipse :)
The current code does require you to know the Gist number, so you'll need a web browser to look it up, but we do have search facilities in mind. Also, while the code attempts so, the resulting Gist is not automatically openend in an editor (a bug). Another idea is to just install the egit plugin in Bioclipse :)
What's wrong with just using egit? I think it's pretty stable now.
ReplyDelete